The hepatic expression levels of genes were analyz
Post# of 158196
Quote:
The hepatic expression levels of genes were analyzed by real-time quantitative PCR in morbidly obese patients with normal liver histology (S0, n = 6), in morbidly obese patients with severe steatosis (S3, n = 6), and in morbidly obese patients with severe steatosis and NASH (NASH, n = 6).
https://journals.plos.org/plosone/article?id=...ne.0013577
The patients in Cytodyn's trial were with NASH.
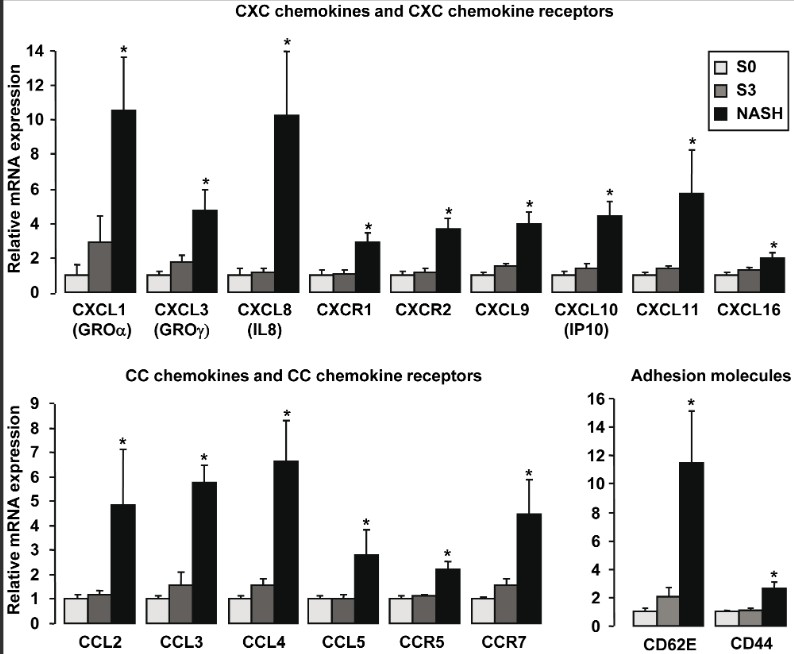
Quote:
CCR5 seems to be upregulated as the disease progresses, (in what I tend to think of as an exponential manner, which means, that the closer the patient gets to Cirrhosis
Who would have ever guessed that.
Quote:
The 700mg haplotype did better than the 700mg group over all. But the outcomes are based on a difference between baseline and end of trial. One would assuredly see a greater difference with a CCR5 blocker if there's more CCR5.
But why would 350mg be higher than not only the entire 700mg group but also the 700mg haplotype group? In the paper baseline levels of cT1 and PDFF in 700mg were not broken out like they were for 350mg. Which could be because of very similar baselines in 700mg that were lower than for 350mg. Remember the p value is based on the difference between baseline and end of trial. With lower disease severity you would of course have a smaller difference and that could be why 350mg beat 700mg. It would also explain the vast difference in chemistry and serum markers between 350mg and 700mg.
https://investorshangout.com/post/view?id=6427476
 (2)
(2) (0)
(0)