Sean Harrison did his own analysis and write-up on
Post# of 157863

https://twitter.com/Sean_Harrison2/status/114...8913644544
https://seanharrisonblog.com/2019/06/20/ccr5-...ans-is-it/
Quote:
Conclusion
This paper has been enormously successful, at least in terms of publicity. I also like to think that my “post-publication peer review” and Rasmus’s reply represents a nice collaborative exchange that wouldn’t have been possible without Twitter. I suppose I could have sent an email, but that doesn’t feel as useful somehow.
However, there are many flaws with the paper that should have been addressed in peer review. I’d love to ask the reviewers why they didn’t insist on the following:
The sample should be well defined, i.e. “white British ancestry” not “British ancestry”
Standard exclusions should be made for sex mismatches, sex chromosome aneuploidy, participants with outliers in heterozygosity and missing rates, and withdrawals from the study (this is important to mention in all papers, right?)
Relatedness should either be accounted for in the analysis (e.g. Bolt-LMM) or related participants should be removed
Population stratification should be both addressed in the analysis (maximum principal components and centre) and the limitations
All effect estimates should have confidence intervals (I mean, come on)
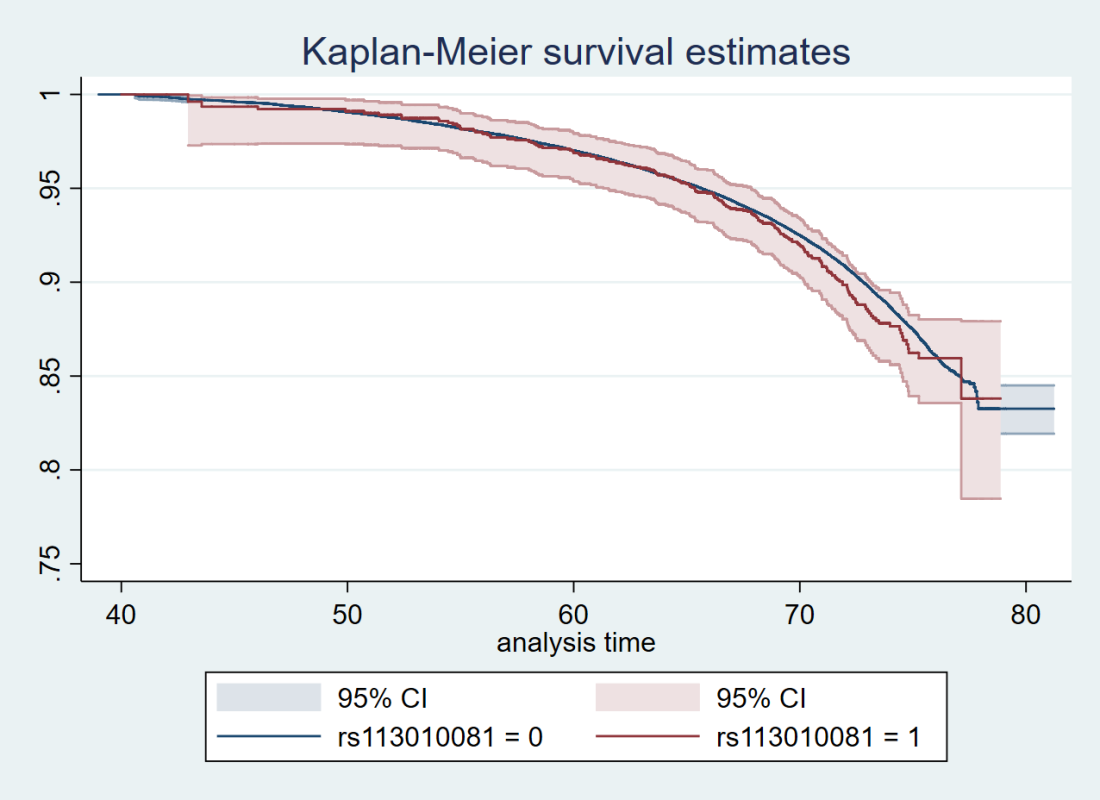
All survival curves should have confidence intervals (ditto)
If it’s a survival analysis, surely Cox regression is better than ratios of survival rates? Also, somewhere it would be useful to note how many people died, and separately for each dosage
One-tailed P values need a huge prior belief to be used in preference to two-tailed P values
Over-reliance on P values in interpretation of results is also to be avoided
Choice of SNP, if you’re only using one SNP, is super important. If your SNP has a very different minor allele frequency from a published paper using a very similar dataset, maybe reference it and state why that might be. Also note if there is any missing data, and why that might be ok
When there is an online supplement to a published paper, I see no legitimate reason why “data not shown” should ever appear
Putting code online is wonderful. Indeed, the paper has a good amount of transparency, with code put on github, and lab notes also put online. I really like this.
So, do I believe “CCR5-∆32 is deleterious in the homozygous state in humans”?
No, I don’t believe there is enough evidence to say that the delta-32 deletion in CCR-5 affects mortality in people of white British ancestry, let alone people of other ancestries.
I know that this post has likely come out far too late to dam the flood of news articles that have already come out. But I kind of hope that what I’ve done will be useful to someone.
 (1)
(1) (0)
(0)